Throughout my research journey, I've collaborated with various universities and research centers. These partnerships have afforded me enriching experiences, allowing me to delve into plant breeding across a variety of crops, including dry beans, corn, sugarcane, and cowpea. My engagements have honed my skills, enabling me to work proficiently in both field and laboratory environments and to develop expertise in bioinformatics and data analysis. At MSU, my academic coursework and hands-on research, combined with collaborative projects, have deepened my understanding of plant breeding. Furthermore, I've had the privilege of working alongside colleagues from diverse research groups spanning the United States, Canada, Colombia, Rwanda, Uganda, and Zambia. These interactions with individuals from varied cultural, academic, and professional backgrounds have been invaluable. You can explore more about my past and current work below.
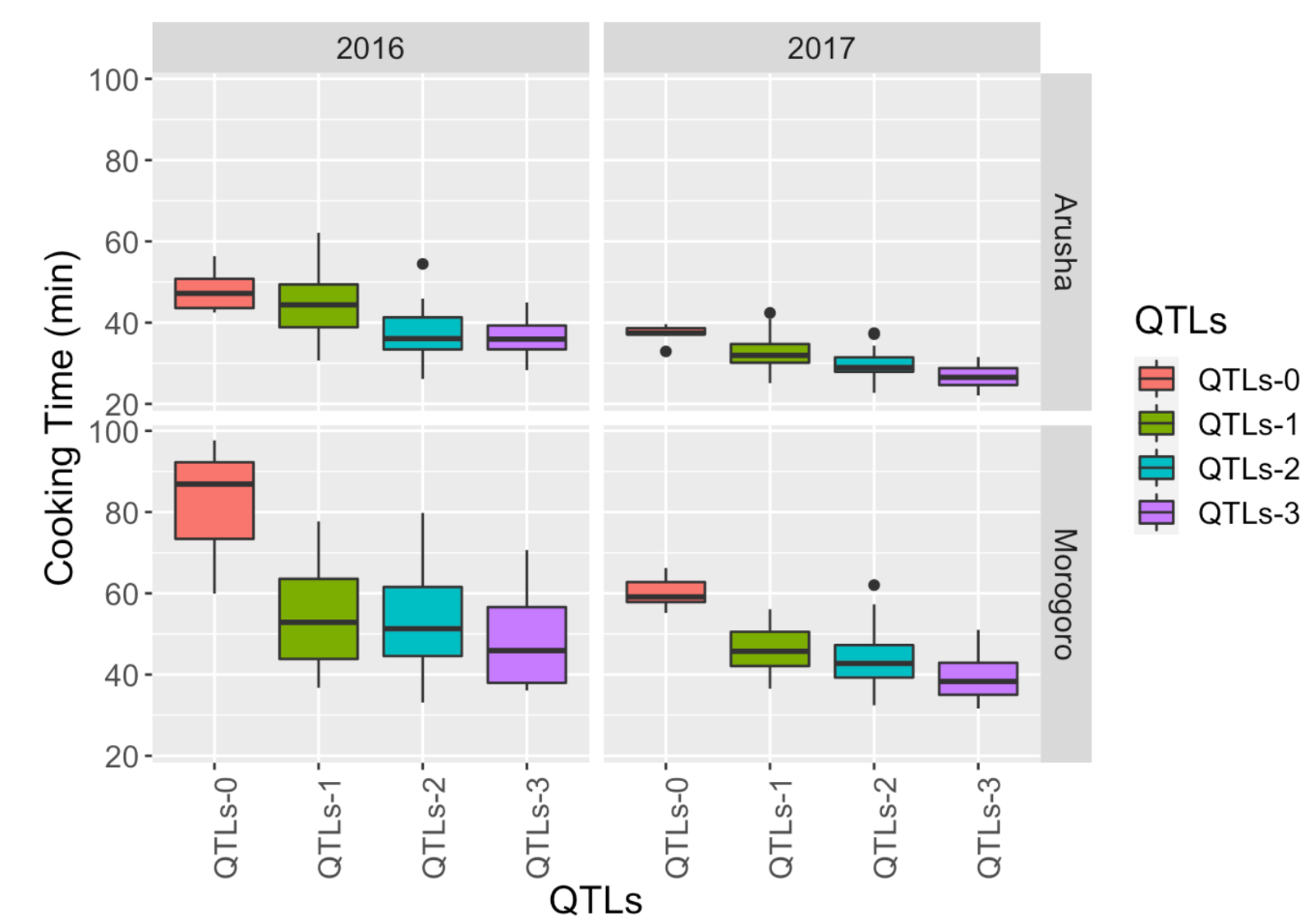
Cooking time significantly influences consumer preferences for dry beans. In our study, we genotyped a biparental population planted across Africa and the United States. We identified three QTLs that not only reduce cooking time but also enhance the protein content in the seeds.
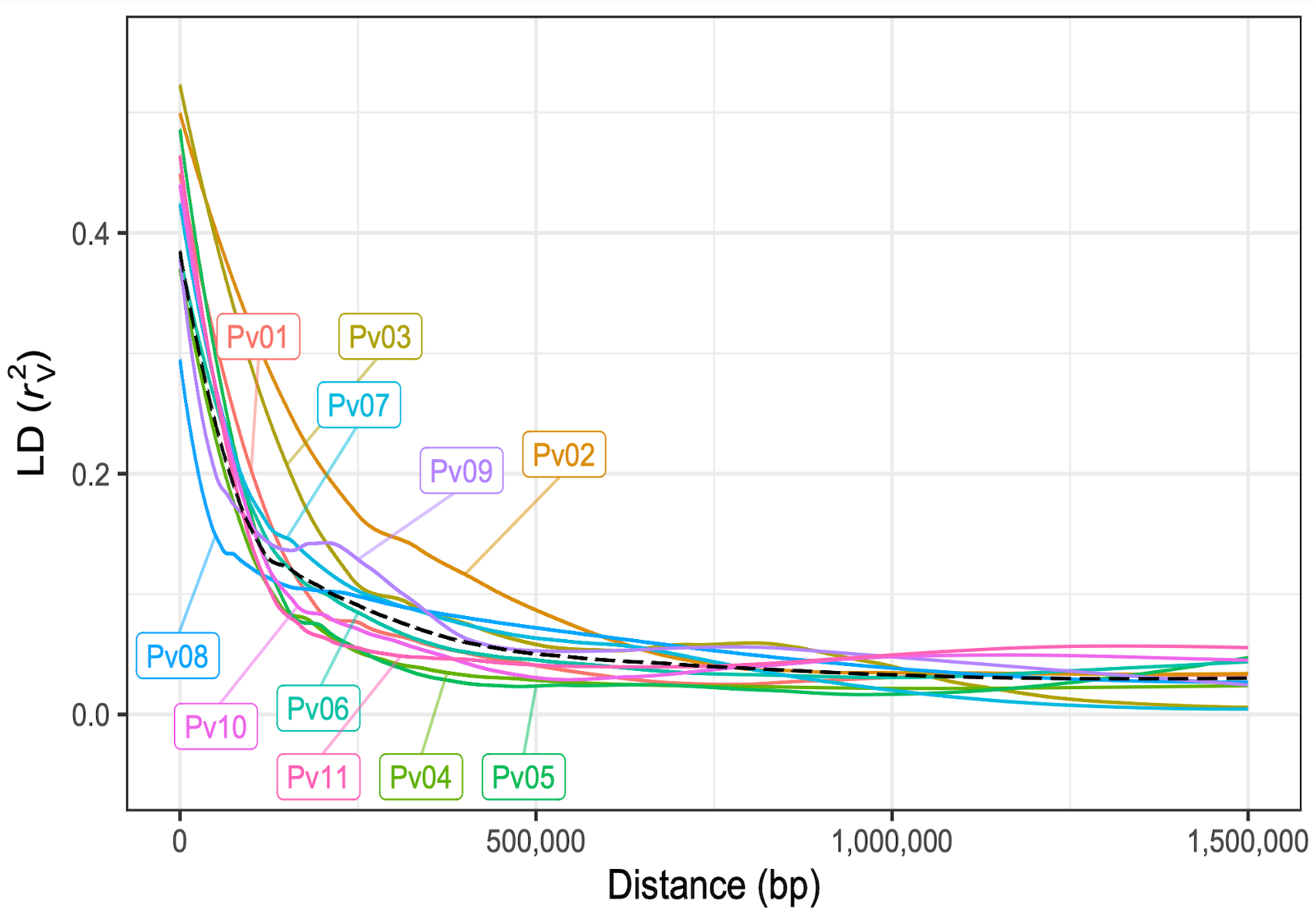
Using a MAGIC population, we explored the genetic underpinnings of phenotypic responses to drought in dry beans. The genotypic and phenotypic data collected led to the identification of promising candidate genes to use in plant breeding.
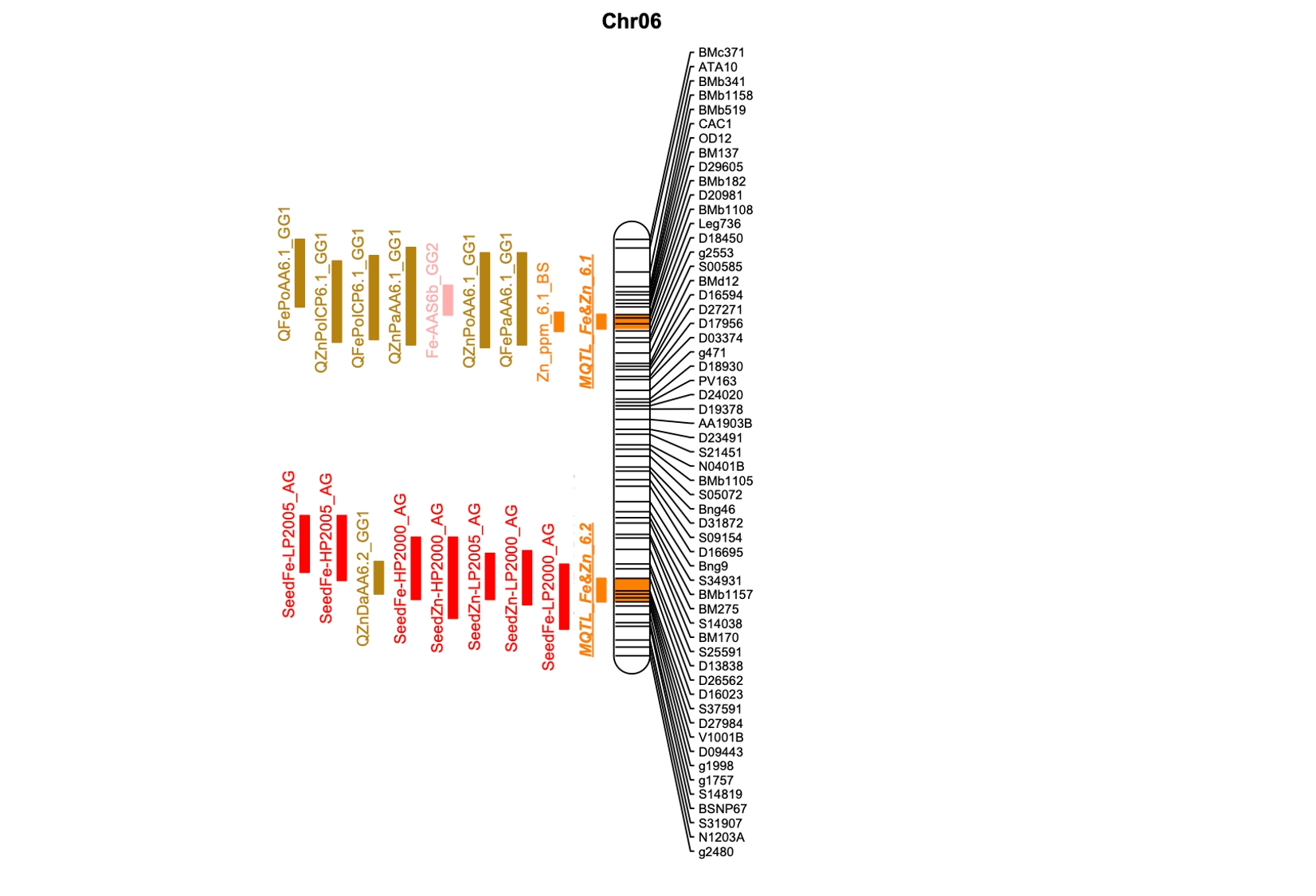
Dry beans are crucial in human nutrition, serving as a significant source of microelements. In our research, we conducted a meta-analysis across seven populations to identify genomic regions associated with the concentration of Fe and Zn in seeds. The meta-QTL regions identified hold promise for marker-assisted selection aimed at enhancing the Fe and Zn concentrations in dry beans.
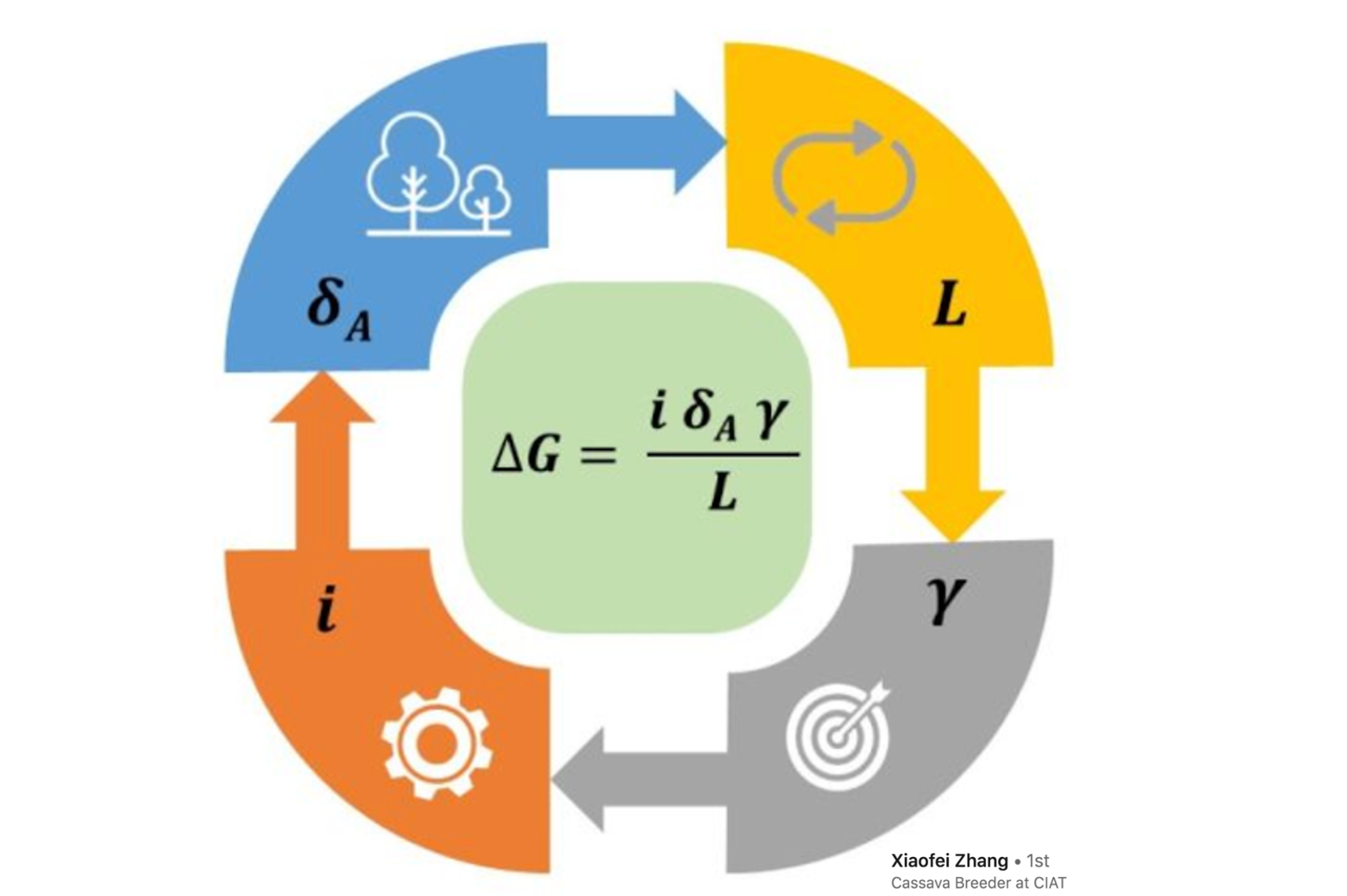
In a diverse panel of dry beans, we applied genomic selection to target end-use quality traits. We observed particularly high prediction accuracies for extreme phenotypes. These results suggest that genomic selection can effectively optimize germplasm selection, with the goals of increasing zinc and protein concentrations in the grain and shortening cooking time.
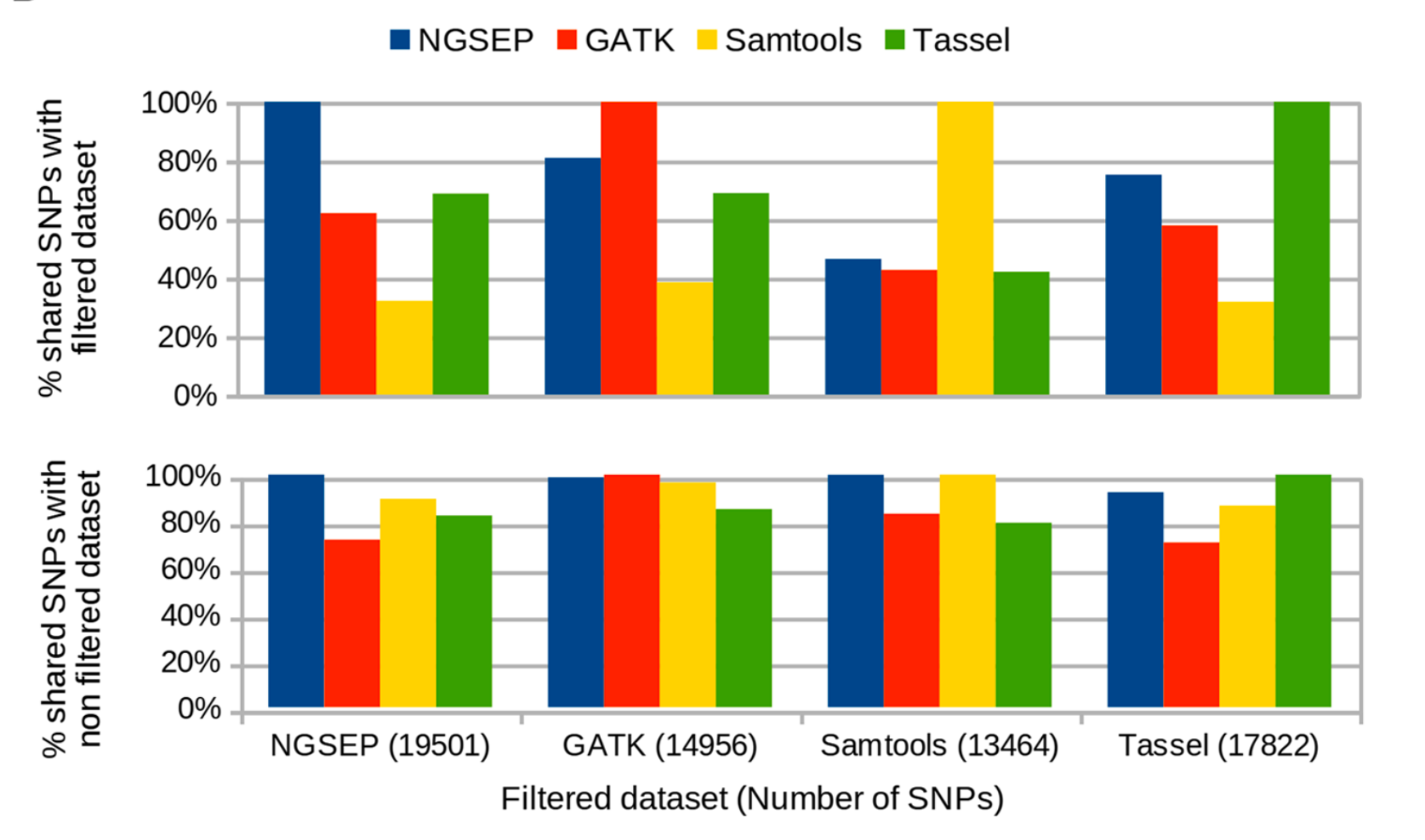
NGSEP stands out as a robust, precise, and efficient bioinformatics software for analyzing high-throughput sequencing data.